Work Package 2: Risk Prediction
The vision for WP2 is to develop an operational risk model that gives an individual diabetes risk prediction for the entire population of Denmark, Greenland and the Faroe Islands. We will achieve this by designing a risk prediction algorithm based exclusively on existing register-based data. WP2 will have a duration of 3 years (Years 1 to 3 of the project) and includes partners from all 7 Steno Centers.
Protocol
The most up-to-date version of the full WP2 analysis protocol(1) is published online on Zenodo.
Methods
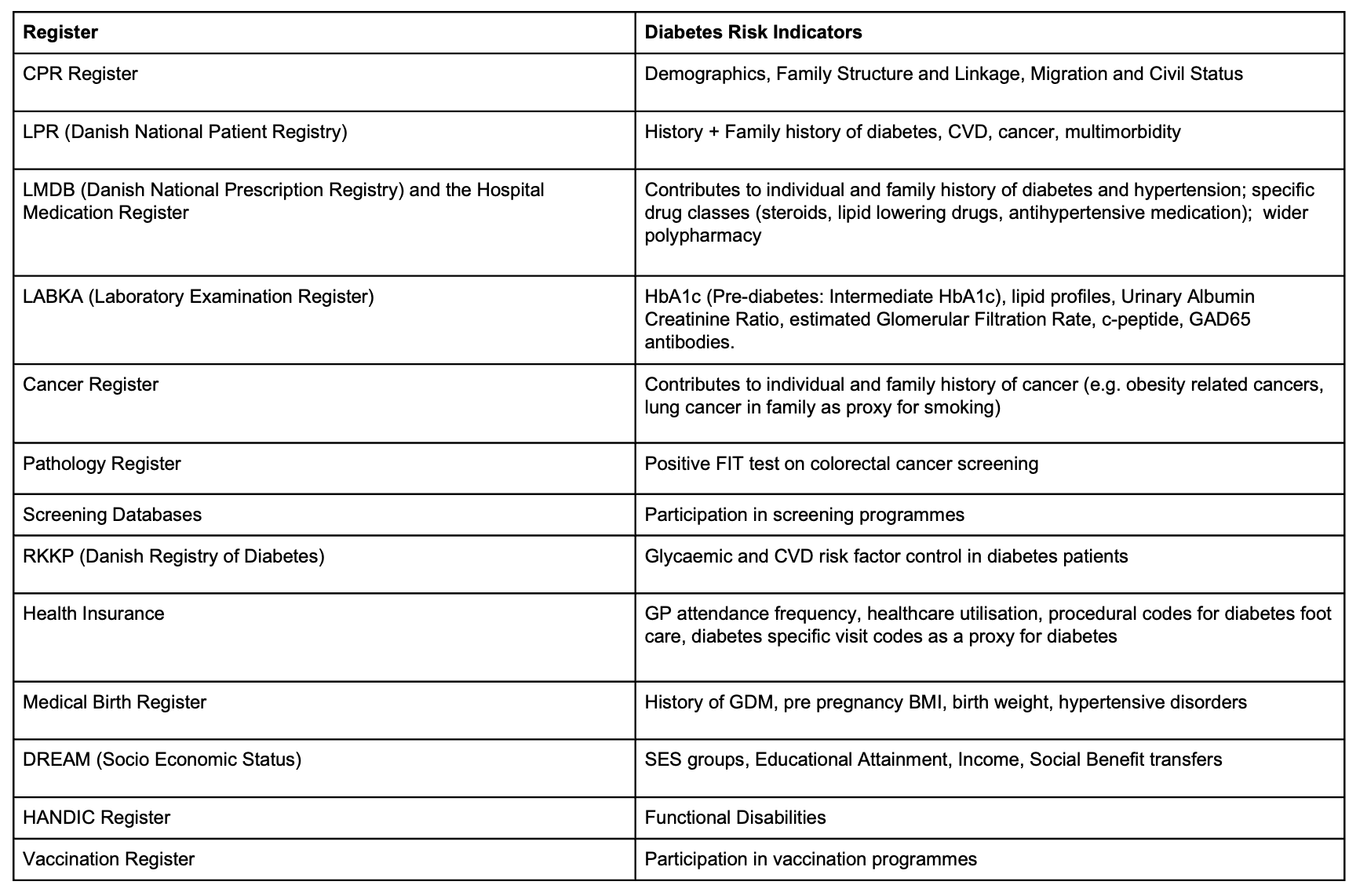
The risk model will be developed using Danish nationwide register data in the DST environment (Figure 1).
Models will then be transferred and adapted to clinical settings by application in EMR environments in Denmark, Greenland and the Faroe Islands.
Analysis will be hosted within the existing DST project database at SDCA where large register projects are already in place, comprising all updated versions of the Danish registers needed for the execution of the project. The outcome, incident diabetes will be derived from various registers according to the Open Source Diabetes Classifier developed at SDCA, which is extensively documented and updated.
We will develop, compare and combine two different approaches: Modular: a traditional statistical approach (Poisson models) applied in parallel within different domains (e.g. GDM/reproductive health, medication history, diagnostic history, socio-economic status), followed by an ensemble model which produces a yearly updated summary risk prediction. Machine Learning (ML): data driven ML methods will be applied to identify informative risk indicators and high-level interactions across all domains, and a risk prediction model will be developed using methods adapted to handle time-to-event data. e.g. survival versions of random forests, XGBoost, DeepCox, DeepHit, Dynamic-DeepHit, MultiSurv. Model interpretability will be an important consideration, in order to quantify the source of the estimated risk both at the population level (HR, variable importance) and at the individual level (SHAP values).
Greenlandic and Faroese data is clinically more comprehensive than Danish register data, making direct inclusion of EMR data in the development and transfer of model(s) necessary and feasible in these two countries. Diabetes in Greenland has a larger genetic component and differs in other dimensions that may help identify unique risk factors. Performance of the model will be assessed using C-statistics. Internal validation will be performed using bootstrapping and k-fold cross-validation, and/or internal-external cross validation. External validation of the model will be done in similar register data from e.g. Sweden or Scotland. The model will be reported according to the TRIPOD+ AI standards.
Several smaller or larger models can be developed based on data accessibility and practical usability requirements. Models will be integrated into the ongoing SAMBLIK project, which will provide secure cross-sectorial insight into individual-level data across the primary and secondary care sectors, taking in data from municipalities and regions. The ambition is for the WP2 risk model(s) to become an integrated module in the SAMBLIK Diabetes tab, thus giving clinicians direct individual insight into the trajectory of diabetes risk and contributing risk factors. At the group level municipalities will be able to use SAMBLIK for a detailed local socio economic and geographic map of diabetes risk, e.g. for use in WP4 activities.
Risk groups identified in WP2 will inform the focus and structure of WP4 and will be used as a benchmark to examine the additional benefit of deep phenotyping in WP3.